Intake I - find, browse and access intake-esm collections#
Overview
🎯 objectives: Learn how to use intake to find, browse and access intake-esm ESM-collections
⌛ time_estimation: “30min”
☑️ requirements: intake_esm.__version__ >= 2023.4.*, at least 10GB memory.
© contributors: k204210
⚖ license:
Agenda
In this part, you learn
Motivation of intake-esm
Features of intake and intake-esm
Browse through catalogs
Data access via intake-esm
We follow here the guidance presented by intake-esm on its repository.
Motivation of intake-esm#
Simulations of the Earth’s climate and weather generate huge amounts of data. These data are often persisted on different storages in a variety of formats (netCDF, zarr, etc…). Finding, investigating, loading these data assets into compute-ready data containers costs time and effort. The data user needs to know what data sets are available, the attributes describing each data set, before loading a specific data set and analyzing it.
Intake-esmaddresses these issues by providing necessary functionality for searching, discovering, data access and data loading.
For intake users, many data preparation tasks are no longer necessary. They do not need to know:
🌍 where data is saved
🪧 how data is saved
📤 how data should be loaded
but still can search, discover, access and load data of a project.
Features of intake and intake-esm#
Intake is a generic cataloging system for listing data sources. As a plugin, intake-esm is built on top of intake, pandas, and xarray and configures intake such that it is able to also load and process ESM data.
display catalogs as clearly structured tables 📄 inside jupyter notebooks for easy investigation
browse 🔍 through the catalog and select your data without
being next to the data (e.g. logged in on dkrz’s luv)
knowing the project’s data reference syntax i.e. the storage tree hierarchy and path and file name templates
open climate data in an analysis ready dictionary of
xarraydatasets 🎁
All required information for searching, accessing and loading the catalog’s data is configured within the catalogs:
🌍 where data is saved
users can browse data without knowing the data storage platform including e.g. the root path of the project and the directory syntax
data of different platforms (cloud or disk) can be combined in one catalog
on mid term, intake catalogs can be a single point of access
🪧 how data is saved
users can work with a xarray dataset representation of the data no matter whether it is saved in grb, netcdf or zarr format.
catalogs can contain more information an therefore more search facets than obvious from names and pathes of the data.
📤 how data should be loaded
users work with an aggregated xarray dataset representation which merges files/assets perfectly fitted to the project’s data model design.
with xarray and the underlying dask library, data which are larger than the RAM can be loaded
In this tutorial, we load a CMIP6 catalog which contains all data from the pool on DKRZ’s mistral disk storage. CMIP6 is the 6th phase of the Coupled Model Intercomparison Project and builds the data base used in the IPCC AR6. The CMIP6 catalog contains all data that is published or replicated at the ESGF node at DKRZ.
Terminology: Catalog, Catalog file and Collection#
We align our wording with intake’s glossary which is still evolving. The names overlap with other definitions, making it difficult to keep track. Here we try to give an overview of the hierarchy of catalog terms:
a top level catalog file 📋 is the main catalog of an institution which will be opened first. It contains other project catalogs 📖 📖 📖. Such catalogs can be assigned an intake driver which is used to open and load the catalog within the top level catalog file. Technically, a catalog file 📋 is
is a
.yamlfilecan be opened with
open_catalog, e.g.:
intake.open_catalog(["https://dkrz.de/s/intake"])
intake drivers also named plugins are specified for catalogs becaues they load specific data sets. There are many driver libraries for intake. .
a catalog 📖 (or collection) is defined by two parts:
a description of a group of data sets. It describes how to load assets of the data set(s) with the specified driver. This group forms an entity. E.g., all CMIP6 data sets can be collected in a catalog.
a collection of all assets of the data set(s).
the collection can be included in the catalog or separately saved in a data base 🗂. In the latter case, the catalog references the data base, e.g.:
"catalog_file": "/mnt/lustre02/work/ik1017/Catalogs/dkrz_cmip6_disk.csv.gz"
Note
The term collection is often used synonymically for catalog.
a intake-esm catalog 📖 consists of a
.jsonfile (the description) and the underlying data base. The data base is either provided within the.jsonfile or as a.csv.gzformatted list.
The intake-esm catalog can be opened with intake-esm’s function intake.open_esm_datastore() where the .json part is the argument, e.g:
intake.open_esm_datastore("https://gitlab.dkrz.de/data-infrastructure-services/intake-esm/-/raw/master/esm-collections/cloud-access/dkrz_cmip6_disk.json")
#note that intake_esm is imported with `import intake` as a plugin
import intake
#to find out the version of intake-esm, you can do:
import intake_esm
intake_esm.__version__
'2023.6.14'
Open and browse through catalogs#
intake (not intake-esm) opens catalog-files in yaml format. These contain information about additonal sources: other catalogs/collections which will be loaded with specific plugins/drivers. The command is open_catalog.
You only need to remember one URL as the single point of access for DKRZ’s intake catalogs: The DKRZ top level catalog can be accessed via dkrz.de/s/intake . Intake will only follow this redirect if a specific parser is activated. This can be done by providing the url in a list.
#dkrz_catalog=intake.open_catalog(["https://dkrz.de/s/intake"])
#dkrz_catalog=intake.open_catalog(["/pool/data/Catalogs/dkrz_catalog.yaml"])
#
#only for the web page we need to take the original link:
dkrz_catalog=intake.open_catalog(["https://gitlab.dkrz.de/data-infrastructure-services/intake-esm/-/raw/intake2022/esm-collections/cloud-access/dkrz_catalog.yaml"])
c6=dkrz_catalog["dkrz_cmip6_disk"]
Note
Right now, two versions of the top level catalog file exist: One for accessing the catalog via cloud, one for via disk. They however contain the same content.
We can look into the catalog with print and list
Over the time, many collections have been created. dkrz_catalog is a main catalog prepared to keep an overview of all other collections. list shows all sub project catalogs which are available at DKRZ.
list(dkrz_catalog)
['dkrz_cmip5_archive',
'dkrz_cmip5_disk',
'dkrz_cmip6_cloud',
'dkrz_cmip6_disk',
'dkrz_cordex_disk',
'dkrz_dyamond-winter_disk',
'dkrz_era5_disk',
'dkrz_monsoon_disk',
'dkrz_mpige_disk',
'dkrz_nextgems_disk',
'dkrz_palmod2_disk']
All these catalogs are intake-esm catalogs. You can find this information via the _entries attribute. The line plugin: ['esm_datastore'] refers to intake-esm’s function open_esm_datastore().
print(dkrz_catalog._entries)
{'dkrz_cmip5_archive': name: dkrz_cmip5_archive
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: This is an ESM collection for CMIP5 data accessible on the DKRZ's cera archive
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'product_id', 'institute', 'model', 'experiment', 'frequency', 'modeling_realm', 'mip_table', 'ensemble_member', 'version', 'variable', 'temporal_subset', 'uri', 'format', 'jblob_file']
obj: {{CATALOG_DIR}}/dkrz_cmip5_archive.json, 'dkrz_cmip5_disk': name: dkrz_cmip5_disk
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: This is an ESM collection for CMIP5 data accessible on the DKRZ's Lustre disk storage system in /work/kd0956/CMIP5/data/cmip5/
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'product_id', 'institute', 'model', 'experiment', 'frequency', 'modeling_realm', 'mip_table', 'ensemble_member', 'version', 'variable', 'temporal_subset', 'uri', 'format']
obj: {{CATALOG_DIR}}/dkrz_cmip5_disk.json, 'dkrz_cmip6_cloud': name: dkrz_cmip6_cloud
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: This is an ESM collection for CMIP6 data accessible on DKRZ's swift cloud store
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'activity_id', 'source_id', 'institution_id', 'experiment_id', 'member_id', 'table_id', 'variable_id', 'grid_label', 'version', 'uri', 'format']
obj: {{CATALOG_DIR}}/dkrz_cmip6_cloud.json, 'dkrz_cmip6_disk': name: dkrz_cmip6_disk
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: This is a ESM-collection for CMIP6 data on DKRZ's disk storage system
direct_access: forbid
user_parameters: [{'name': 'additional_cmip6_disk_columns', 'description': None, 'type': 'list[str]', 'default': ['units', 'path', 'opendap_url', 'long_name']}, {'name': 'additional_era5_disk_columns', 'description': None, 'type': 'list[str]', 'default': ['path', 'units', 'long_name', 'step', 'short_name']}, {'name': 'cataloonie_columns', 'description': None, 'type': 'list[str]', 'default': ['project', 'institution_id', 'source_id', 'experiment_id', 'simulation_id', 'realm', 'frequency', 'time_reduction', 'grid_label', 'grid_id', 'level_type', 'time_min', 'time_max', 'time_range', 'format', 'uri', 'variable_id']}]
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'activity_id', 'source_id', 'institution_id', 'experiment_id', 'member_id', 'dcpp_init_year', 'table_id', 'variable_id', 'grid_label', 'version', 'time_range', 'uri', 'format']
obj: {{CATALOG_DIR}}/dkrz_cmip6_disk.json, 'dkrz_cordex_disk': name: dkrz_cordex_disk
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: This is an ESM collection for CORDEX data accessible on the DKRZ's disk storage system
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'product_id', 'CORDEX_domain', 'institute_id', 'driving_model_id', 'experiment_id', 'member', 'model_id', 'rcm_version_id', 'frequency', 'variable_id', 'version', 'time_range', 'uri', 'format']
obj: {{CATALOG_DIR}}/dkrz_cordex_disk.json, 'dkrz_dyamond-winter_disk': name: dkrz_dyamond-winter_disk
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: Default catalog for ICON-ESM experiments by MPIMet
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'institution_id', 'source_id', 'experiment_id', 'simulation_id', 'realm', 'frequency', 'time_reduction', 'grid_label', 'grid_id', 'level_type', 'time_min', 'time_max', 'time_range', 'format', 'uri', 'variable_id']
obj: {{CATALOG_DIR}}/dkrz_dyamond-winter_disk.json, 'dkrz_era5_disk': name: dkrz_era5_disk
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: This is an ESM collection for ERA5 data accessible on the DKRZ's disk storage system in /work/bk1099/data/
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'era_id', 'dataType', 'level_type', 'frequency', 'stepType', 'table_id', 'code', 'validation_date', 'initialization_date', 'uri', 'format']
obj: {{CATALOG_DIR}}/dkrz_era5_disk.json, 'dkrz_monsoon_disk': name: dkrz_monsoon_disk
container: catalog
plugin: ['yaml_file_cat']
driver: ['yaml_file_cat']
description: Monsoon 2.0
direct_access: forbid
user_parameters: []
metadata:
args:
path: {{CATALOG_DIR}}/dkrz_monsoon_disk.yaml, 'dkrz_mpige_disk': name: dkrz_mpige_disk
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: This is an ESM collection for the Max Planck Institute Grand Ensemble (Maher et al. 2019 https://doi.org/10/gf3kgt) cmorized by CMIP5-standards accessible on the DKRZ's Levante disk storage system in /work/mh1007/CMOR/MPI-GE
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['product_id', 'institute', 'model', 'experiment', 'frequency', 'modeling_realm', 'mip_table', 'ensemble_member', 'variable', 'temporal_subset', 'version', 'uri', 'format']
obj: {{CATALOG_DIR}}/dkrz_mpige_disk.json, 'dkrz_nextgems_disk': name: dkrz_nextgems_disk
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: Default catalog for ICON-ESM experiments by MPIMet
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'institution_id', 'source_id', 'experiment_id', 'simulation_id', 'realm', 'frequency', 'time_reduction', 'grid_label', 'grid_id', 'level_type', 'time_min', 'time_max', 'time_range', 'format', 'uri', 'variable_id']
obj: {{CATALOG_DIR}}/dkrz_nextgems_disk.json, 'dkrz_palmod2_disk': name: dkrz_palmod2_disk
container: xarray
plugin: ['esm_datastore']
driver: ['esm_datastore']
description: This is a ESM-collection for Palmod2 data on DKRZ's disk storage system
direct_access: forbid
user_parameters: []
metadata:
args:
read_csv_kwargs:
usecols: ['project', 'source_id', 'institution_id', 'experiment_id', 'member_id', 'table_id', 'variable_id', 'grid_label', 'version', 'format', 'time_range', 'uri']
obj: {{CATALOG_DIR}}/dkrz_palmod2_disk.json}
The DKRZ ESM-Collections follow a name template:
dkrz_${project}_${store}[_${auxiliary_catalog}]
where
project can be one of the model intercomparison project, e.g.
cmip6,cmip5,cordex,era5ormpi-ge.store is the data store and can be one of:
disk: DKRZ holds a lot of data on a consortial disk space on the file system of the High Performance Computer (HPC) where it is accessible for every HPC user. Working next to the data on the file system will be the fastest way possible.cloud: A small subset is transferred into DKRZ’s cloud in order to test the performance. swift is DKRZ’s cloud storage.archive: A lot of data exists in the band archive of DKRZ. Before it can be accessed, it has to be retrieved. Therefore, catalogs forhsmare limited in functionality but still convenient for data browsing.
auxiliary_catalog can be grid
Why that convention?:
dkrz: Assume you work with internation collections. Than it may become important that you know from where the data comes, e.g. if only pathes on a local file system are given as the locations of the data.
project: Project’s data standards differ from each other so that different catalog attributes are required to identify a single asset in a project data base.
store: Intake-esm cannot load data from all stores. Before data from the archive can be accessed, it has to be retrieved. Therefore, the opening function is not working for catalog merged for all stores.
Best practice for naming catalogs:
Use small letters for all values
Do NOT use
_as a separator in valuesDo not repeat values of other attributes (“dkrz_dkrz-dyamond”)
We could directly start to work with two intake catalog at the same time.
Let’s have a look into a master catalog of Pangeo:
pangeo=intake.open_catalog("https://raw.githubusercontent.com/pangeo-data/pangeo-datastore/master/intake-catalogs/master.yaml")
pangeo
master:
args:
path: https://raw.githubusercontent.com/pangeo-data/pangeo-datastore/master/intake-catalogs/master.yaml
description: Pangeo Master Data Catalog
driver: intake.catalog.local.YAMLFileCatalog
metadata: {}
list(pangeo)
['ocean', 'atmosphere', 'climate', 'hydro']
While DKRZ’s master catalog has one sublevel, Pangeo’s is a nested one. We can access another yaml catalog which is also a parent catalog by simply:
pangeo.climate
climate:
args:
path: https://raw.githubusercontent.com/pangeo-data/pangeo-datastore/master/intake-catalogs/climate.yaml
description: Pangeo Climate Dataset Catalog. Include model ensembles such as CMIP6
and LENS.
driver: intake.catalog.local.YAMLFileCatalog
metadata:
catalog_dir: https://raw.githubusercontent.com/pangeo-data/pangeo-datastore/master/intake-catalogs
Pangeo’s ESM collections are one level deeper in the catalog tree:
list(pangeo.climate)
['cmip6_gcs',
'cmip6_s3',
'GFDL_CM2_6',
'GFDL_CM2_6_s3',
'tracmip',
'tracmip_s3']
The intake-esm catalogs#
We now look into a catalog which is opened by the plugin intake-esm.
An ESM (Earth System Model) collection file is a
JSONfile that conforms to the ESM Collection Specification. When provided a link/path to an esm collection file, intake-esm establishes a link to a database (CSVfile) that contains data assets locations and associated metadata (i.e., which experiment, model, the come from).
Since the data base of the CMIP6 ESM Collection is about 100MB in compressed format, it takes up to a minute to load the catalog.
Note
The project catalogs contain only valid and current project data. They are constantly updated.
If your work is based on a catalog and a subset of the data from it, be sure to save that subset so you can later compare your database to the most current catalog.
esm_col=dkrz_catalog.dkrz_cmip6_disk
print(esm_col)
</work/ik1017/Catalogs/dkrz_cmip6_disk catalog with 17246 dataset(s) from 6132163 asset(s)>
intake-esm gives us an overview over the content of the ESM collection. The ESM collection is a data base described by specific attributes which are technically columns. Each project data standard is the basis for the columns and used to parse information given by the path and file names.
The pure display of esm_col shows us the number of unique values in each column. Since each uri refers to one file, we can conclude that the DKRZ-CMIP6 ESM Collection contains 6.1 Mio Files in 2022.
The data base is loaded into an underlying pandas dataframe which we can access with esm_col.df. esm_col.df.head() displays the first rows of the table:
esm_col.df.head()
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | dcpp_init_year | version | time_range | project | format | uri | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | AerChemMIP | BCC | BCC-ESM1 | hist-piAer | r1i1p1f1 | AERmon | c2h6 | gn | NaN | v20200511 | 185001-201412 | CMIP6 | netcdf | /work/ik1017/CMIP6/data/CMIP6/AerChemMIP/BCC/B... |
| 1 | AerChemMIP | BCC | BCC-ESM1 | hist-piAer | r1i1p1f1 | AERmon | c2h6 | gn | NaN | v20200511 | 185001-201412.nc.modified | CMIP6 | netcdf | /work/ik1017/CMIP6/data/CMIP6/AerChemMIP/BCC/B... |
| 2 | AerChemMIP | BCC | BCC-ESM1 | hist-piAer | r1i1p1f1 | AERmon | c3h6 | gn | NaN | v20200511 | 185001-201412 | CMIP6 | netcdf | /work/ik1017/CMIP6/data/CMIP6/AerChemMIP/BCC/B... |
| 3 | AerChemMIP | BCC | BCC-ESM1 | hist-piAer | r1i1p1f1 | AERmon | c3h8 | gn | NaN | v20200511 | 185001-201412 | CMIP6 | netcdf | /work/ik1017/CMIP6/data/CMIP6/AerChemMIP/BCC/B... |
| 4 | AerChemMIP | BCC | BCC-ESM1 | hist-piAer | r1i1p1f1 | AERmon | cdnc | gn | NaN | v20200522 | 185001-201412 | CMIP6 | netcdf | /work/ik1017/CMIP6/data/CMIP6/AerChemMIP/BCC/B... |
We can find out details about esm_col with the object’s attributes. esm_col.esmcol_data contains all information given in the JSON file. We can also focus on some specific attributes.
dict(esm_col.esmcat)["description"]
"This is a ESM-collection for CMIP6 data on DKRZ's disk storage system which will be loaded from a source file which is in the cloud (see catalog_file)"
print("What is this catalog about? \n" + dict(esm_col.esmcat)["description"])
#
print("The link to the data base: "+ dict(esm_col.esmcat)["catalog_file"])
What is this catalog about?
This is a ESM-collection for CMIP6 data on DKRZ's disk storage system which will be loaded from a source file which is in the cloud (see catalog_file)
The link to the data base: https://swift.dkrz.de/v1/dkrz_a44962e3ba914c309a7421573a6949a6/intake-esm/dkrz_cmip6_disk.csv.gz
Advanced: To find out how many datasets are available, we can use pandas functions (drop columns that are irrelevant for a dataset, drop the duplicates, keep one):
cat = esm_col.df.drop(['uri','time_range'],axis=1).drop_duplicates(keep="first")
print(len(cat))
1355084
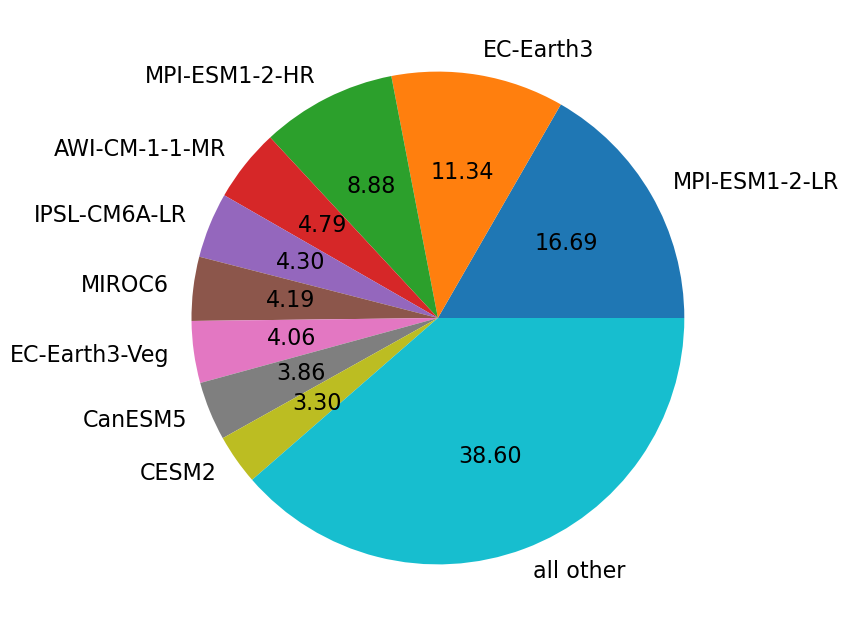
def pieplot(gbyelem) :
#groupby, sort and select the ten largest
global c6
size = c6.df.groupby([gbyelem]).size().sort_values(ascending=False)
size10 = size.nlargest(10)
#Sum all others as 10th entry
size10[9] = sum(size[9:])
size10.rename(index={size10.index.values[9]:'all other'},inplace=True)
#return a pie plot
return size10.plot.pie(figsize=(18,8),ylabel='',autopct='%.2f', fontsize=16)
pieplot("source_id")
<Axes: >
Browse through the data of the ESM collection#
You will browse the collection technically by setting values the column names of the underlying table. Per default, the catalog was loaded with all cmip6 attributes/columns that define the CMIP6 data standard:
esm_col.df.columns
Index(['activity_id', 'institution_id', 'source_id', 'experiment_id',
'member_id', 'table_id', 'variable_id', 'grid_label', 'dcpp_init_year',
'version', 'time_range', 'project', 'format', 'uri'],
dtype='object')
These are configured in the top level catalog so you do not need to open the catalog to see the columns
dkrz_catalog._entries["dkrz_cmip6_disk"]._open_args
{'read_csv_kwargs': {'usecols': ['project',
'activity_id',
'source_id',
'institution_id',
'experiment_id',
'member_id',
'dcpp_init_year',
'table_id',
'variable_id',
'grid_label',
'version',
'time_range',
'uri',
'format']},
'obj': '{{CATALOG_DIR}}/dkrz_cmip6_disk.json'}
Most of the time, we want to set more than one attribute for a search. Therefore, we define a query dictionary and use the search function of the esm_col object. In the following case, we look for temperature at surface in monthly resolution for 3 different experiments:
query = dict(
variable_id="tas",
table_id="Amon",
experiment_id=["piControl", "historical", "ssp370"])
# piControl = pre-industrial control, simulation to represent a stable climate from 1850 for >100 years.
# historical = historical Simulation, 1850-2014
# ssp370 = Shared Socioeconomic Pathways (SSPs) are scenarios of projected socioeconomic global changes. Simulation covers 2015-2100
cat = esm_col.search(**query)
cat
/work/ik1017/Catalogs/dkrz_cmip6_disk catalog with 165 dataset(s) from 17427 asset(s):
| unique | |
|---|---|
| activity_id | 3 |
| institution_id | 32 |
| source_id | 65 |
| experiment_id | 3 |
| member_id | 205 |
| table_id | 1 |
| variable_id | 1 |
| grid_label | 3 |
| dcpp_init_year | 0 |
| version | 204 |
| time_range | 1798 |
| project | 1 |
| format | 1 |
| uri | 17427 |
| derived_variable_id | 0 |
We could also use Wildcards. For example, in order to find out which ESMs of the institution MPI-M have produced data for our subset:
cat.search(source_id="MPI-ES*")
/work/ik1017/Catalogs/dkrz_cmip6_disk catalog with 9 dataset(s) from 1180 asset(s):
| unique | |
|---|---|
| activity_id | 2 |
| institution_id | 3 |
| source_id | 3 |
| experiment_id | 3 |
| member_id | 31 |
| table_id | 1 |
| variable_id | 1 |
| grid_label | 1 |
| dcpp_init_year | 0 |
| version | 8 |
| time_range | 162 |
| project | 1 |
| format | 1 |
| uri | 1180 |
| derived_variable_id | 0 |
We can find out which models have submitted data for at least one of them by:
cat.unique()["source_id"]
['BCC-ESM1',
'TaiESM1',
'AWI-CM-1-1-MR',
'AWI-ESM-1-1-LR',
'BCC-CSM2-MR',
'CAMS-CSM1-0',
'CAS-ESM2-0',
'FGOALS-f3-L',
'FGOALS-g3',
'IITM-ESM',
'CanESM5',
'CanESM5-CanOE',
'CMCC-CM2-HR4',
'CMCC-CM2-SR5',
'CMCC-ESM2',
'CNRM-CM6-1',
'CNRM-CM6-1-HR',
'CNRM-ESM2-1',
'ACCESS-ESM1-5',
'ACCESS-CM2',
'E3SM-1-0',
'E3SM-1-1',
'E3SM-1-1-ECA',
'EC-Earth3',
'EC-Earth3-AerChem',
'EC-Earth3-CC',
'EC-Earth3-LR',
'EC-Earth3-Veg',
'EC-Earth3-Veg-LR',
'FIO-ESM-2-0',
'MPI-ESM-1-2-HAM',
'INM-CM4-8',
'INM-CM5-0',
'IPSL-CM5A2-INCA',
'IPSL-CM6A-LR',
'KIOST-ESM',
'MIROC-ES2H',
'MIROC-ES2L',
'MIROC6',
'HadGEM3-GC31-LL',
'HadGEM3-GC31-MM',
'UKESM1-0-LL',
'ICON-ESM-LR',
'MPI-ESM1-2-HR',
'MPI-ESM1-2-LR',
'MRI-ESM2-0',
'GISS-E2-1-G',
'GISS-E2-1-G-CC',
'GISS-E2-1-H',
'GISS-E2-2-G',
'CESM2',
'CESM2-FV2',
'CESM2-WACCM',
'CESM2-WACCM-FV2',
'NorCPM1',
'NorESM1-F',
'NorESM2-LM',
'NorESM2-MM',
'KACE-1-0-G',
'GFDL-CM4',
'GFDL-ESM4',
'NESM3',
'SAM0-UNICON',
'CIESM',
'MCM-UA-1-0']
If we instead look for the models that have submitted data for ALL experiments, we use the require_all_on keyword argument:
cat = esm_col.search(require_all_on=["source_id"], **query)
cat.unique()["source_id"]
['ACCESS-CM2',
'ACCESS-ESM1-5',
'AWI-CM-1-1-MR',
'BCC-CSM2-MR',
'BCC-ESM1',
'CAMS-CSM1-0',
'CESM2',
'CESM2-WACCM',
'CMCC-CM2-SR5',
'CMCC-ESM2',
'CNRM-CM6-1',
'CNRM-CM6-1-HR',
'CNRM-ESM2-1',
'CanESM5',
'CanESM5-CanOE',
'EC-Earth3',
'EC-Earth3-AerChem',
'EC-Earth3-Veg',
'EC-Earth3-Veg-LR',
'FGOALS-f3-L',
'FGOALS-g3',
'GFDL-ESM4',
'GISS-E2-1-G',
'IITM-ESM',
'INM-CM4-8',
'INM-CM5-0',
'IPSL-CM6A-LR',
'KACE-1-0-G',
'MCM-UA-1-0',
'MIROC-ES2L',
'MIROC6',
'MPI-ESM-1-2-HAM',
'MPI-ESM1-2-HR',
'MPI-ESM1-2-LR',
'MRI-ESM2-0',
'NorESM2-LM',
'NorESM2-MM',
'TaiESM1',
'UKESM1-0-LL']
Note that only the combination of a variable_id and a table_id is unique in CMIP6. If you search for tas in all tables, you will find many entries more:
query = dict(
variable_id="tas",
# table_id="Amon",
experiment_id=["piControl", "historical", "ssp370"])
cat = esm_col.search(**query)
cat.unique()["table_id"]
['Amon',
'day',
'3hr',
'6hrPlev',
'6hrPlevPt',
'AERhr',
'CFsubhr',
'ImonAnt',
'ImonGre']
Be careful when you search for specific time slices. Each frequency is connected with a individual name template for the filename. If the data is yearly, you have YYYY-YYYY whereas you have YYYYMM-YYYYMM for monthly data.
How to load more columns#
Intake allows to load only a subset of the columns that is inside the intake-esm catalog. Since the memory usage of intake-esm is high, the default columns are only a subset from all possible columns. Sometimes, other columns are of interest:
If you work remotely away from the data, you can use the opendap_url’s to access the subset of interest for all files published at DKRZ. The opendap_url is an additional column that can also be loaded.
We can define 3 different column name types for the usage of intake catalogs:
Default attributes which are loaded from the main catalog and which can be seen via
_entries[CATNAME]._open_args.Overall attributes or template attributes which should be defined for ALL catalogs at DKRZ (exceptions excluded). At DKRZ, we use the newly defined Cataloonie scheme template which can be found via
dkrz_catalog.metadata["parameters"]["cataloonie_columns"]. With these template attributes, there may be redundancy in the columns. They exist to simplify merging catalogs across projects.Additional attributes which are not necessary to identify a single asset but helpful for users. You can find these via
dkrz_catalog.metadata["parameters"]["additional_PROJECT_columns"]
So, for CMIP6 there are:
dkrz_catalog.metadata["parameters"]["additional_cmip6_disk_columns"]
{'default': ['units', 'path', 'opendap_url', 'long_name'], 'type': 'list[str]'}
Tip
You may find variable_ids in the catalog which are not obvious or abbrevations for a clear variable name. In that cases you would need additional information like a long_name of the variable. For CMIP6, we provided the catalog with this long_name so you could add it as a column.
So, this is the instruction how to open the catalog with additional columns:
create a combination of all your required columns:
cols=dkrz_catalog._entries["dkrz_cmip6_disk"]._open_args["read_csv_kwargs"]["usecols"]+["opendap_url"]
open the dkrz_cmip6_disk catalog with the
csv_kwargskeyword argument in this way:
#esm_col=dkrz_catalog.dkrz_cmip6_disk(read_csv_kwargs=dict(usecols=cols))
⭐ The customization of catalog columns allows highest flexibility for intake users.
⭐ In theory, we could add many more columns with additional information because ot all have to be loaded from the data base.
Warning
The number of columns determines the required memory.
Tip
If you work from remote and also want to access the data remotely, load the opendap_url column.
Access and load data of the ESM collection#
With the power of xarray, intake can load your subset into a dictionary of datasets. We therefore focus on the data of MPI-ESM1-2-LR:
#case insensitive?
query = dict(
variable_id="tas",
table_id="Amon",
source_id="MPI-ESM1-2-LR",
experiment_id="historical")
cat = esm_col.search(**query)
cat
/work/ik1017/Catalogs/dkrz_cmip6_disk catalog with 1 dataset(s) from 279 asset(s):
| unique | |
|---|---|
| activity_id | 1 |
| institution_id | 1 |
| source_id | 1 |
| experiment_id | 1 |
| member_id | 31 |
| table_id | 1 |
| variable_id | 1 |
| grid_label | 1 |
| dcpp_init_year | 0 |
| version | 3 |
| time_range | 9 |
| project | 1 |
| format | 1 |
| uri | 279 |
| derived_variable_id | 0 |
You can find out which column intake uses to access the data via the following keyword:
cat.esmcat.assets.column_name
'uri'
As we are working with the _disk catalog, uri contains pathes to the files on filesystem. If you are working from remote, you would have
to change the catalog’s attribute
path_column_nameto opendap_url.to reassign the
formatcolumn from netcdf to opendap
as follows:
#cat.esmcat.assets.column_name="opendap_url"
#newdf=cat.df.copy()
#newdf.loc[:,"format"]="opendap"
#cat.df=newdf
Intake-ESM natively supports the following data formats or access formats (since opendap is not really a file format):
netcdf
opendap
zarr
You can also open grb data but right now only by specifying xarray’s attribute engine in the open function which is defined in the following. I.e., it does not make a difference if you specify grb as format.
You can find an example in the era5 notebook.
The function to open data is to_dataset_dict.
We recommend to set a keyword argument xarray_open_kwargs for the chunk size of the variable’s data array. Otherwise, xarray may choose too large chunks. Most often, your data contains a time dimension so that you could set xarray_open_kwargs={"chunks":{"time":1}}.
If your collection contains zarr formatted data, you need to add another keyword argument zarr_kwargs.
Unfortunately, this has changed in new versions:
- DEPRICATED The trick is: You can just specify both. Intake knows from the `format` column which *kwargs* should be taken.
help(cat.to_dataset_dict)
Help on method to_dataset_dict in module intake_esm.core:
to_dataset_dict(xarray_open_kwargs: dict[str, typing.Any] = None, xarray_combine_by_coords_kwargs: dict[str, typing.Any] = None, preprocess: Callable = None, storage_options: dict[pydantic.types.StrictStr, typing.Any] = None, progressbar: pydantic.types.StrictBool = None, aggregate: pydantic.types.StrictBool = None, skip_on_error: pydantic.types.StrictBool = False, **kwargs) -> dict[str, xarray.core.dataset.Dataset] method of intake_esm.core.esm_datastore instance
Load catalog entries into a dictionary of xarray datasets.
Column values, dataset keys and requested variables are added as global
attributes on the returned datasets. The names of these attributes can be
customized with :py:class:`intake_esm.utils.set_options`.
Parameters
----------
xarray_open_kwargs : dict
Keyword arguments to pass to :py:func:`~xarray.open_dataset` function
xarray_combine_by_coords_kwargs: : dict
Keyword arguments to pass to :py:func:`~xarray.combine_by_coords` function.
preprocess : callable, optional
If provided, call this function on each dataset prior to aggregation.
storage_options : dict, optional
fsspec Parameters passed to the backend file-system such as Google Cloud Storage,
Amazon Web Service S3.
progressbar : bool
If True, will print a progress bar to standard error (stderr)
when loading assets into :py:class:`~xarray.Dataset`.
aggregate : bool, optional
If False, no aggregation will be done.
skip_on_error : bool, optional
If True, skip datasets that cannot be loaded and/or variables we are unable to derive.
Returns
-------
dsets : dict
A dictionary of xarray :py:class:`~xarray.Dataset`.
Examples
--------
>>> import intake
>>> cat = intake.open_esm_datastore("glade-cmip6.json")
>>> sub_cat = cat.search(
... source_id=["BCC-CSM2-MR", "CNRM-CM6-1", "CNRM-ESM2-1"],
... experiment_id=["historical", "ssp585"],
... variable_id="pr",
... table_id="Amon",
... grid_label="gn",
... )
>>> dsets = sub_cat.to_dataset_dict()
>>> dsets.keys()
dict_keys(['CMIP.BCC.BCC-CSM2-MR.historical.Amon.gn', 'ScenarioMIP.BCC.BCC-CSM2-MR.ssp585.Amon.gn'])
>>> dsets["CMIP.BCC.BCC-CSM2-MR.historical.Amon.gn"]
<xarray.Dataset>
Dimensions: (bnds: 2, lat: 160, lon: 320, member_id: 3, time: 1980)
Coordinates:
* lon (lon) float64 0.0 1.125 2.25 3.375 ... 355.5 356.6 357.8 358.9
* lat (lat) float64 -89.14 -88.03 -86.91 -85.79 ... 86.91 88.03 89.14
* time (time) object 1850-01-16 12:00:00 ... 2014-12-16 12:00:00
* member_id (member_id) <U8 'r1i1p1f1' 'r2i1p1f1' 'r3i1p1f1'
Dimensions without coordinates: bnds
Data variables:
lat_bnds (lat, bnds) float64 dask.array<chunksize=(160, 2), meta=np.ndarray>
lon_bnds (lon, bnds) float64 dask.array<chunksize=(320, 2), meta=np.ndarray>
time_bnds (time, bnds) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
pr (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 600, 160, 320), meta=np.ndarray>
xr_dict = cat.to_dataset_dict(
#xarray_open_kwargs=
#dict(
# chunks=dict(
# time=60
# ),
#consolidated=True
#decode_times=True,
#use_cftime=True
#)
)
xr_dict
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.source_id.experiment_id.table_id.grid_label'
{'CMIP.MPI-ESM1-2-LR.historical.Amon.gn': <xarray.Dataset>
Dimensions: (time: 1980, bnds: 2, lat: 96, lon: 192, member_id: 31,
dcpp_init_year: 1)
Coordinates:
* time (time) datetime64[ns] 1850-01-16T12:00:00 ... 2014-12-16T...
time_bnds (time, bnds) datetime64[ns] dask.array<chunksize=(240, 2), meta=np.ndarray>
* lat (lat) float64 -88.57 -86.72 -84.86 ... 84.86 86.72 88.57
lat_bnds (lat, bnds) float64 dask.array<chunksize=(96, 2), meta=np.ndarray>
* lon (lon) float64 0.0 1.875 3.75 5.625 ... 354.4 356.2 358.1
lon_bnds (lon, bnds) float64 dask.array<chunksize=(192, 2), meta=np.ndarray>
height float64 2.0
* member_id (member_id) object 'r10i1p1f1' 'r11i1p1f1' ... 'r9i1p1f1'
* dcpp_init_year (dcpp_init_year) float64 nan
Dimensions without coordinates: bnds
Data variables:
tas (member_id, dcpp_init_year, time, lat, lon) float32 dask.array<chunksize=(1, 1, 240, 96, 192), meta=np.ndarray>
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
contact: cmip6-mpi-esm@dkrz.de
data_specs_version: 01.00.30
... ...
intake_esm_attrs:grid_label: gn
intake_esm_attrs:project: CMIP6
intake_esm_attrs:_data_format_: netcdf
CDO: Climate Data Operators version 2.0.0rc2...
variant_info: Branched from the PMIP past2k experimen...
intake_esm_dataset_key: CMIP.MPI-ESM1-2-LR.historical.Amon.gn}
Intake was able to aggregate many files into only one dataset:
The
time_rangecolumn was used to concat data along thetimedimensionThe
member_idcolumn was used to generate a new dimension
The underlying dask package will only load the data into memory if needed. Note that attributes which disagree from file to file, e.g. tracking_id, are excluded from the dataset.
How intake-esm should open and aggregate the assets is configured in the aggregation_control part of the description:
cat.esmcat.aggregation_control.aggregations
[Aggregation(type=<AggregationType.union: 'union'>, attribute_name='variable_id', options={}),
Aggregation(type=<AggregationType.join_existing: 'join_existing'>, attribute_name='time_range', options={'compat': 'override', 'coords': 'minimal', 'dim': 'time'}),
Aggregation(type=<AggregationType.join_new: 'join_new'>, attribute_name='member_id', options={'compat': 'override', 'coords': 'minimal'}),
Aggregation(type=<AggregationType.join_new: 'join_new'>, attribute_name='dcpp_init_year', options={'compat': 'override', 'coords': 'minimal'})]
Columns can be defined for appending or creating new dimensions. The options are keyword arguments for xarray.
They keys of the dictionary are made with column values defined in the aggregation_control of the intake-esm catalog. These will determine the key_template. The corresponding commands are:
print(cat.esmcat.aggregation_control.groupby_attrs)
#
print(cat.key_template)
['activity_id', 'source_id', 'experiment_id', 'table_id', 'grid_label']
activity_id.source_id.experiment_id.table_id.grid_label
You can work with these keys directly on the intake-esm catalog which will give you an overview over all columns (too long for the web page):
#cat["CMIP.MPI-ESM1-2-HR.historical.Amon.gn"]
If we are only interested in the first dataset of the dictionary, we can pop it out:
xr_dset = xr_dict.popitem()[1]
xr_dset
<xarray.Dataset>
Dimensions: (time: 1980, bnds: 2, lat: 96, lon: 192, member_id: 31,
dcpp_init_year: 1)
Coordinates:
* time (time) datetime64[ns] 1850-01-16T12:00:00 ... 2014-12-16T...
time_bnds (time, bnds) datetime64[ns] dask.array<chunksize=(240, 2), meta=np.ndarray>
* lat (lat) float64 -88.57 -86.72 -84.86 ... 84.86 86.72 88.57
lat_bnds (lat, bnds) float64 dask.array<chunksize=(96, 2), meta=np.ndarray>
* lon (lon) float64 0.0 1.875 3.75 5.625 ... 354.4 356.2 358.1
lon_bnds (lon, bnds) float64 dask.array<chunksize=(192, 2), meta=np.ndarray>
height float64 2.0
* member_id (member_id) object 'r10i1p1f1' 'r11i1p1f1' ... 'r9i1p1f1'
* dcpp_init_year (dcpp_init_year) float64 nan
Dimensions without coordinates: bnds
Data variables:
tas (member_id, dcpp_init_year, time, lat, lon) float32 dask.array<chunksize=(1, 1, 240, 96, 192), meta=np.ndarray>
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
contact: cmip6-mpi-esm@dkrz.de
data_specs_version: 01.00.30
... ...
intake_esm_attrs:grid_label: gn
intake_esm_attrs:project: CMIP6
intake_esm_attrs:_data_format_: netcdf
CDO: Climate Data Operators version 2.0.0rc2...
variant_info: Branched from the PMIP past2k experimen...
intake_esm_dataset_key: CMIP.MPI-ESM1-2-LR.historical.Amon.gnTroubleshooting#
The variables are collected in one dataset. This requires that the dimensions and coordinates must be the same over all files. Otherwise, xarray cannot merge these together.
For CMIP6, most of the variables collected in one table_id should be on the same dimensions and coordinates. Unfortunately, there are exceptions.:
a few variables are requested for time slices only.
sometimes models use different dimension names from file to file
Using the preprocessing keyword argument can help to rename dimensions before merging.
For Intake providers: the more information on the dimensions and coordinates provided already in the catalog, the better the aggregation control.
Pangeo’s data store#
Let’s have a look into Pangeo’s ESM Collection as well. This is accessible via cloud from everywhere - you only need internet to load data. We use the same query as in the example before.
pangeo.climate.discover()
{'container': 'catalog',
'shape': None,
'dtype': None,
'metadata': {'catalog_dir': 'https://raw.githubusercontent.com/pangeo-data/pangeo-datastore/master/intake-catalogs'}}
import intake
pangeo_cmip6=intake.open_esm_datastore("https://storage.googleapis.com/cmip6/pangeo-cmip6.json")
cat = pangeo_cmip6.search(**query)
cat
pangeo-cmip6 catalog with 1 dataset(s) from 10 asset(s):
| unique | |
|---|---|
| activity_id | 1 |
| institution_id | 1 |
| source_id | 1 |
| experiment_id | 1 |
| member_id | 10 |
| table_id | 1 |
| variable_id | 1 |
| grid_label | 1 |
| zstore | 10 |
| dcpp_init_year | 0 |
| version | 1 |
| derived_variable_id | 0 |
There are differences between the collections because
Pangeo provides files in consolidated,
zarrformatted datasets which correspond tozstoreentries in the catalog instead ofpaths oropendap_urls.The
zarrdatasets are already aggregated over time so there is no need for atime_rangecolumn
If we now open the data with intake, we have to specify keyword arguments as follows:
dset_dict = cat.to_dataset_dict(
zarr_kwargs={"consolidated": True}#, "decode_times": True, "use_cftime": True}
)
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
/tmp/ipykernel_614/1732716172.py:1: DeprecationWarning: cdf_kwargs and zarr_kwargs are deprecated and will be removed in a future version. Please use xarray_open_kwargs instead.
dset_dict = cat.to_dataset_dict(
dset_dict
{'CMIP.MPI-M.MPI-ESM1-2-LR.historical.Amon.gn': <xarray.Dataset>
Dimensions: (lat: 96, bnds: 2, lon: 192, member_id: 10,
dcpp_init_year: 1, time: 1980)
Coordinates:
height float64 2.0
* lat (lat) float64 -88.57 -86.72 -84.86 ... 84.86 86.72 88.57
lat_bnds (lat, bnds) float64 dask.array<chunksize=(96, 2), meta=np.ndarray>
* lon (lon) float64 0.0 1.875 3.75 5.625 ... 354.4 356.2 358.1
lon_bnds (lon, bnds) float64 dask.array<chunksize=(192, 2), meta=np.ndarray>
* time (time) datetime64[ns] 1850-01-16T12:00:00 ... 2014-12-16T...
time_bnds (time, bnds) datetime64[ns] dask.array<chunksize=(1980, 2), meta=np.ndarray>
* member_id (member_id) object 'r10i1p1f1' 'r1i1p1f1' ... 'r9i1p1f1'
* dcpp_init_year (dcpp_init_year) float64 nan
Dimensions without coordinates: bnds
Data variables:
tas (member_id, dcpp_init_year, time, lat, lon) float32 dask.array<chunksize=(1, 1, 990, 96, 192), meta=np.ndarray>
Attributes: (12/54)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
cmor_version: 3.5.0
contact: cmip6-mpi-esm@dkrz.de
... ...
intake_esm_attrs:variable_id: tas
intake_esm_attrs:grid_label: gn
intake_esm_attrs:version: 20190710
intake_esm_attrs:_data_format_: zarr
DODS_EXTRA.Unlimited_Dimension: time
intake_esm_dataset_key: CMIP.MPI-M.MPI-ESM1-2-LR.historical.Amo...}
dset_dict and xr_dict are the same. You succesfully did the intake tutorial!
Making a quick plot#
The following line exemplifies the ease of the intake’s data processing library chain. On the web page, the interactivity will not work as all plots would have to be loaded which is not feasible.
For more examples, check out the use cases on that web page.
import hvplot.xarray
xr_dset["tas"].squeeze().hvplot.quadmesh(width=600)
See also
This tutorial is part of a series on intake:
You can also do another CMIP6 tutorial from the official intake page.

