Simple visualisation of a CMIP6 dataset with Python Xarray#
this is an adoption of NordicESMhub example to use CMIP6 data stored in the DKRZ data pool
filename = '/work/ik1017/CMIP6/data/CMIP6/CMIP/NCAR/CESM2/historical/r1i1p1f1/Amon/tas/gn/v20190308/tas_Amon_CESM2_historical_r1i1p1f1_gn_185001-201412.nc'
Import python packages#
import xarray as xr
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
import numpy as np
import cftime
ERROR 1: PROJ: proj_create_from_database: Open of /envs/share/proj failed
Open dataset#
Use
xarraypython package to analyze netCDF datasetopen_datasetallows to get all the metadata without loading data into memory.with
xarray, we only load into memory what is needed.
dset = xr.open_dataset(filename, decode_times=True, use_cftime=True)
print(dset)
<xarray.Dataset>
Dimensions: (time: 1980, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
Dimensions without coordinates: nbnd
Data variables:
tas (time, lat, lon) float32 ...
time_bnds (time, nbnd) object ...
lat_bnds (lat, nbnd) float32 ...
lon_bnds (lon, nbnd) float32 ...
Attributes: (12/45)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
case_id: 15
cesm_casename: b.e21.BHIST.f09_g17.CMIP6-historical.001
contact: cesm_cmip6@ucar.edu
creation_date: 2019-01-16T23:34:05Z
... ...
sub_experiment: none
sub_experiment_id: none
branch_time_in_parent: 219000.0
branch_time_in_child: 674885.0
branch_method: standard
further_info_url: https://furtherinfo.es-doc.org/CMIP6.NCAR.CESM2.h...
/envs/lib/python3.11/site-packages/xarray/conventions.py:431: SerializationWarning: variable 'tas' has multiple fill values {1e+20, 1e+20}, decoding all values to NaN.
new_vars[k] = decode_cf_variable(
Get metadata corresponding to near-surface air temperature (tas)#
print(dset['tas'])
<xarray.DataArray 'tas' (time: 1980, lat: 192, lon: 288)>
[109486080 values with dtype=float32]
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 87.17 88.12 89.06 90.0
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
Attributes: (12/19)
cell_measures: area: areacella
cell_methods: area: time: mean
comment: near-surface (usually, 2 meter) air temperature
description: near-surface (usually, 2 meter) air temperature
frequency: mon
id: tas
... ...
time_label: time-mean
time_title: Temporal mean
title: Near-Surface Air Temperature
type: real
units: K
variable_id: tas
dset.time.values
array([cftime.DatetimeNoLeap(1850, 1, 15, 12, 0, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(1850, 2, 14, 0, 0, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(1850, 3, 15, 12, 0, 0, 0, has_year_zero=True),
...,
cftime.DatetimeNoLeap(2014, 10, 15, 12, 0, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(2014, 11, 15, 0, 0, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(2014, 12, 15, 12, 0, 0, 0, has_year_zero=True)],
dtype=object)
Select time#
Select a specific time
dset['tas'].sel(time=cftime.DatetimeNoLeap(1850, 1, 15, 12, 0, 0, 0, 2, 15)).plot(cmap = 'coolwarm')
<matplotlib.collections.QuadMesh at 0x7fd228d96dd0>
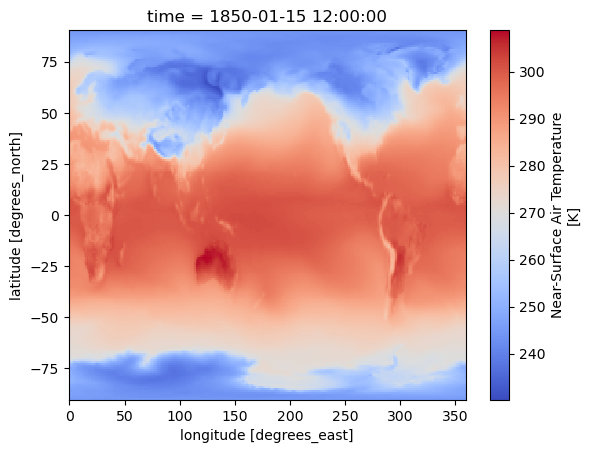
select the nearest time. Here from 1st April 1950
dset['tas'].sel(time=cftime.DatetimeNoLeap(1850, 4, 1), method='nearest').plot(cmap='coolwarm')
<matplotlib.collections.QuadMesh at 0x7fd220492c90>

Customize plot#
Set the size of the figure and add coastlines#
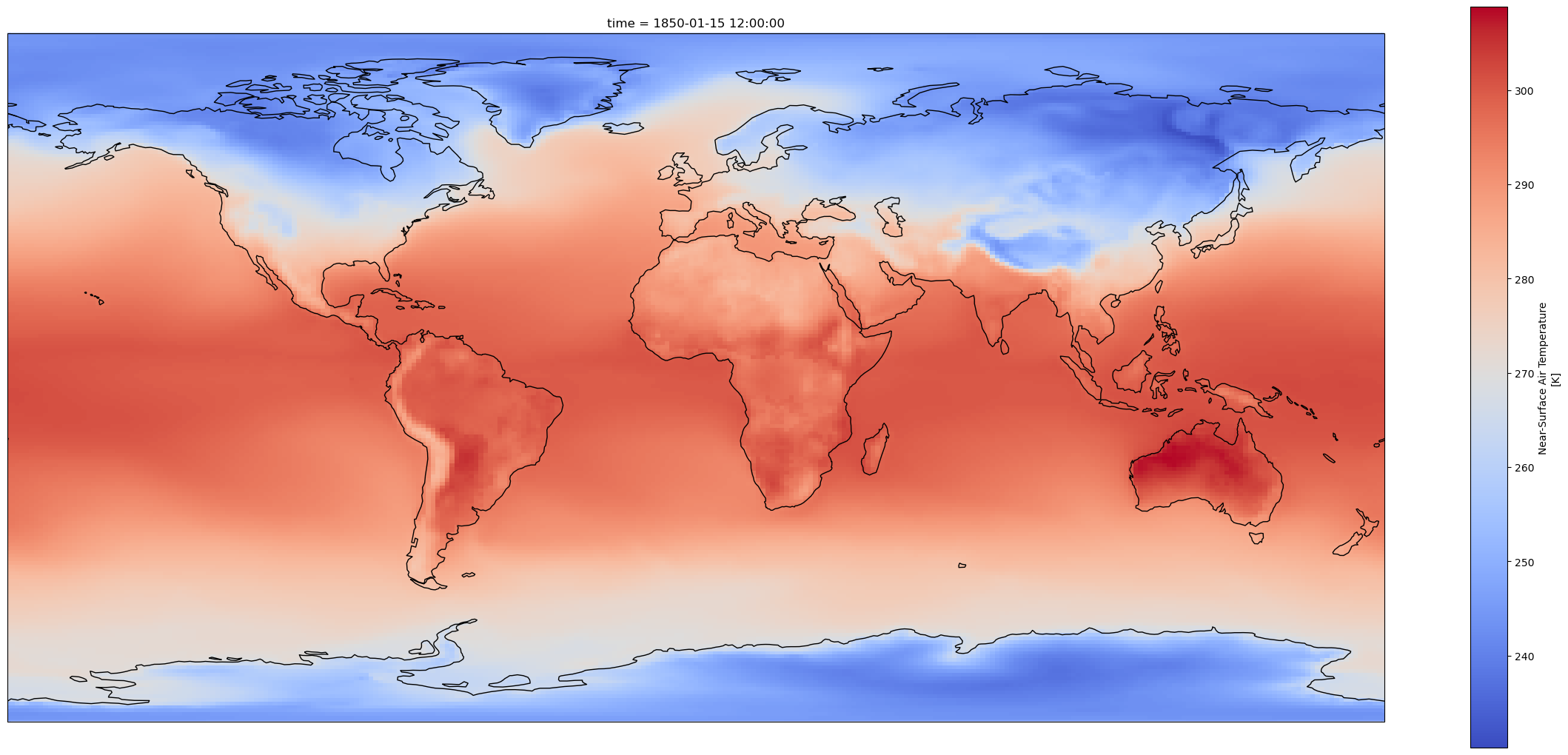
fig = plt.figure(1, figsize=[30,13])
# Set the projection to use for plotting
ax = plt.subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.coastlines()
# Pass ax as an argument when plotting. Here we assume data is in the same coordinate reference system than the projection chosen for plotting
# isel allows to select by indices instead of the time values
dset['tas'].isel(time=0).plot.pcolormesh(ax=ax, cmap='coolwarm')
<cartopy.mpl.geocollection.GeoQuadMesh at 0x7fd228e4f610>
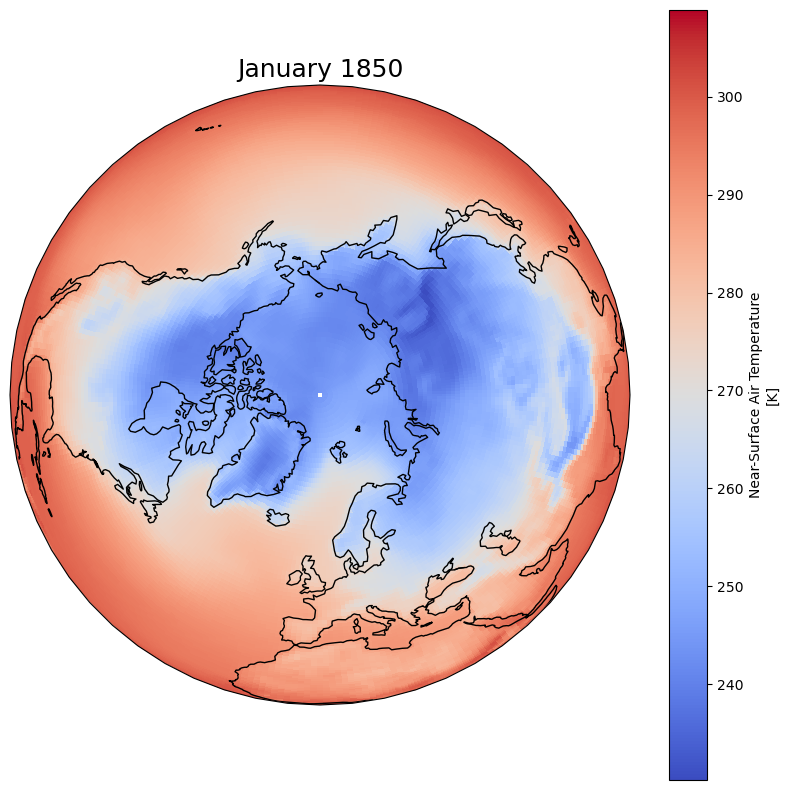
Change plotting projection#
fig = plt.figure(1, figsize=[10,10])
# We're using cartopy and are plotting in Orthographic projection
# (see documentation on cartopy)
ax = plt.subplot(1, 1, 1, projection=ccrs.Orthographic(0, 90))
ax.coastlines()
# We need to project our data to the new Orthographic projection and for this we use `transform`.
# we set the original data projection in transform (here PlateCarree)
dset['tas'].isel(time=0).plot(ax=ax, transform=ccrs.PlateCarree(), cmap='coolwarm')
# One way to customize your title
plt.title(dset.time.values[0].strftime("%B %Y"), fontsize=18)
Text(0.5, 1.0, 'January 1850')
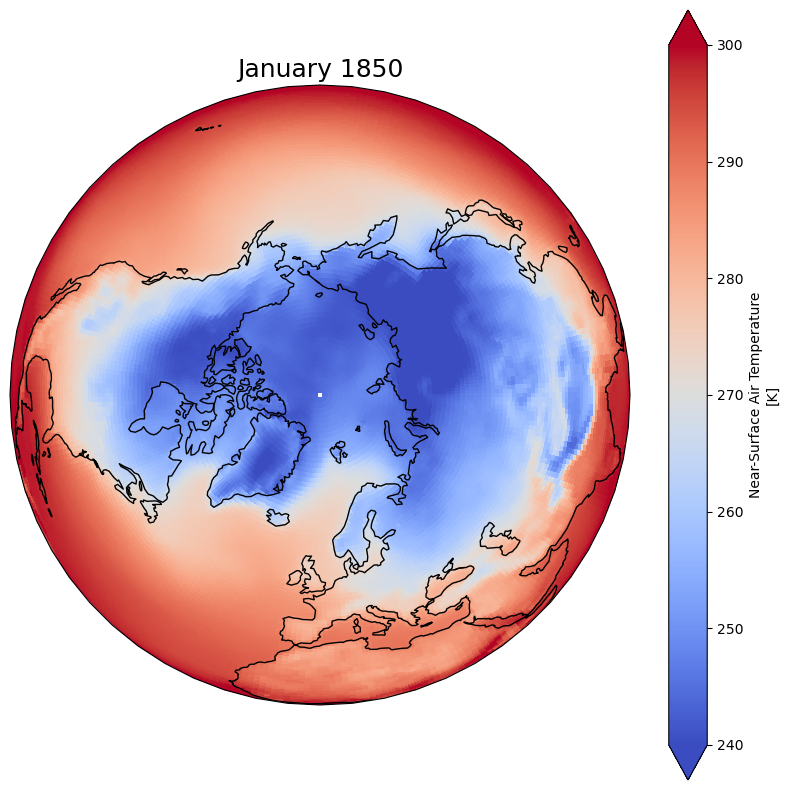
Choose the extent of values#
Fix your minimum and maximum values in your plot and
Use extend so values below the minimum and max
fig = plt.figure(1, figsize=[10,10])
ax = plt.subplot(1, 1, 1, projection=ccrs.Orthographic(0, 90))
ax.coastlines()
# Fix extent
minval = 240
maxval = 300
# pass extent with vmin and vmax parameters
dset['tas'].isel(time=0).plot(ax=ax, vmin=minval, vmax=maxval, transform=ccrs.PlateCarree(), cmap='coolwarm')
# One way to customize your title
plt.title(dset.time.values[0].strftime("%B %Y"), fontsize=18)
Text(0.5, 1.0, 'January 1850')
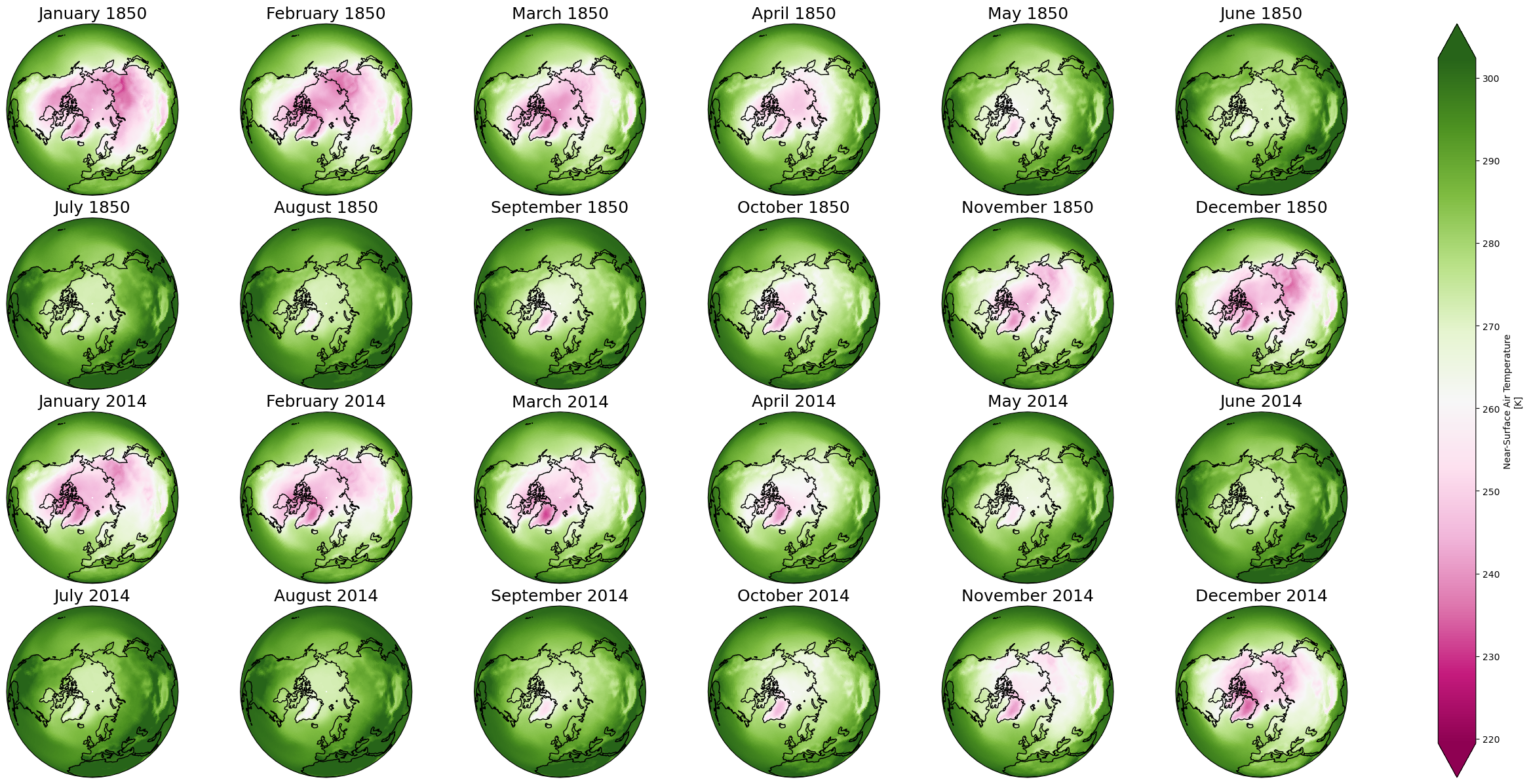
Multiplots#
Faceting#
proj_plot = ccrs.Orthographic(0, 90)
p = dset['tas'].sel(time = dset.time.dt.year.isin([1850, 2014])).plot(x='lon', y='lat',
transform=ccrs.PlateCarree(),
aspect=dset.dims["lon"] / dset.dims["lat"], # for a sensible figsize
subplot_kws={"projection": proj_plot},
col='time', col_wrap=6, robust=True, cmap='PiYG')
# We have to set the map's options on all four axes
for ax,i in zip(p.axes.flat, dset.time.sel(time = dset.time.dt.year.isin([1850, 2014])).values):
ax.coastlines()
ax.set_title(i.strftime("%B %Y"), fontsize=18)
/tmp/ipykernel_1437/1975360723.py:9: DeprecationWarning: self.axes is deprecated since 2022.11 in order to align with matplotlibs plt.subplots, use self.axs instead.
for ax,i in zip(p.axes.flat, dset.time.sel(time = dset.time.dt.year.isin([1850, 2014])).values):
Combine plots with different projections#
fig = plt.figure(1, figsize=[20,10])
# Fix extent
minval = 240
maxval = 300
# Plot 1 for Northern Hemisphere subplot argument (nrows, ncols, nplot)
# here 1 row, 2 columns and 1st plot
ax1 = plt.subplot(1, 2, 1, projection=ccrs.Orthographic(0, 90))
# Plot 2 for Southern Hemisphere
# 2nd plot
ax2 = plt.subplot(1, 2, 2, projection=ccrs.Orthographic(180, -90))
tsel = 0
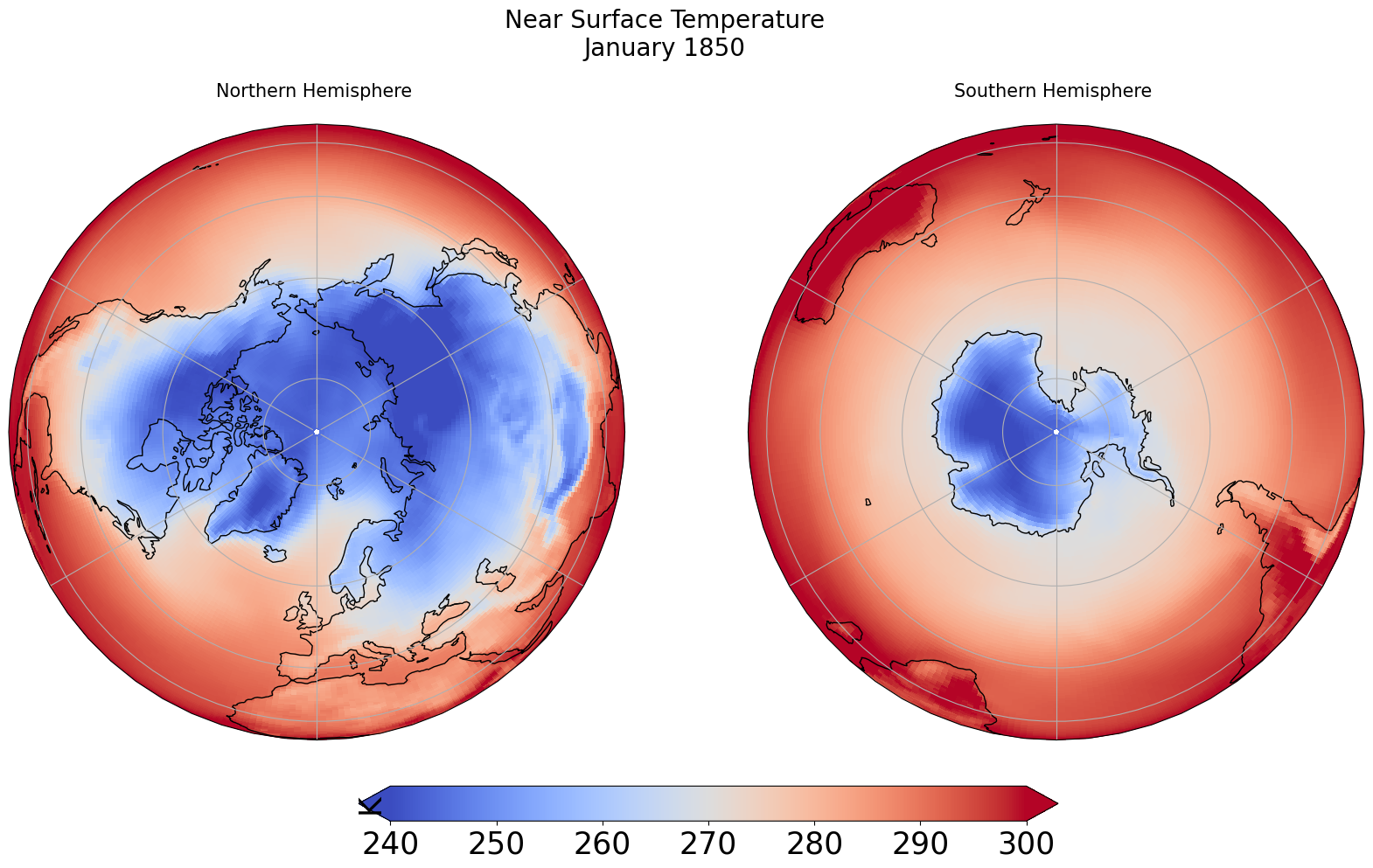
for ax,t in zip([ax1, ax2], ["Northern", "Southern"]):
map = dset['tas'].isel(time=tsel).plot(ax=ax, vmin=minval, vmax=maxval,
transform=ccrs.PlateCarree(),
cmap='coolwarm',
add_colorbar=False)
ax.set_title(t + " Hemisphere \n" , fontsize=15)
ax.coastlines()
ax.gridlines()
# Title for both plots
fig.suptitle('Near Surface Temperature\n' + dset.time.values[tsel].strftime("%B %Y"), fontsize=20)
cb_ax = fig.add_axes([0.325, 0.05, 0.4, 0.04])
cbar = plt.colorbar(map, cax=cb_ax, extend='both', orientation='horizontal', fraction=0.046, pad=0.04)
cbar.ax.tick_params(labelsize=25)
cbar.ax.set_ylabel('K', fontsize=25)
Text(0, 0.5, 'K')